Sélection induite par les plantes des microbes dans la rétroaction de la rhizosphère et des microbes des plantes
Résumé
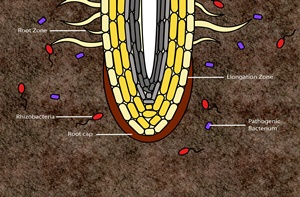
L’effet des plantes sur les microorganismes du sol et les rétroactions entre les microbes et les plantes sont devenus le focus de beaucoup de recherche. Les dernières avancées dans le domaine des interactions plantes-microbes montrent que les plantes sont capables de façonner le microbiome de leur rhizosphère via des mécanismes divers. Dans cette revue, nous rassemblons des résultats de multiples études sur le rôle des plantes dans l’altération de la structure et des fonctions des microorganismes au niveau de la rhizosphère. En outre, nous discutons les rôles de différentes substances phytochimiques dans la médiation de ces effets. Finalement, nous soulignons que l’enrichissement sélectif de microorganismes spécifiques a aussi bien des rétroactions négatives, avec l’accumulation de pathogènes dans la rhizosphère, que positives, suite au recrutement de microflore bénéfique. Une meilleure compréhension des mécanismes qui régissent la sélection de microorganismes ayant des rétroactions positives ouvrira de nouvelles possibilités pour l’amélioration de la production agricole.
Mots-clés: Plantes, microorganismes, rhizosphère, rétroactions.
Téléchargements
Publié-e
Comment citer
Numéro
Rubrique
Licence

Revue Marocaine des Sciences Agronomiques et Vétérinaires est mis à disposition selon les termes de la licence Creative Commons Attribution - Pas d’Utilisation Commerciale - Partage dans les Mêmes Conditions 4.0 International.
Fondé(e) sur une œuvre à www.agrimaroc.org.
Les autorisations au-delà du champ de cette licence peuvent être obtenues à www.agrimaroc.org.

